Tutorial¶
How to make queries on an endpoint like Pathway Commons?¶
BioPAX (Biological Pathway Exchange) is a RDF (Resource Description Framework)/OWL (Web Ontology Language) knowledge representation that can be stored on endpoints in two forms of graphs.
- as Named Graph for each source of data; for example one named graph for Reactome, another for PID, etc.
- or as an unique graph combining all data sources.
A module has been developped to provide a quick description of what users can query on an endpoint in order to forge their queries in consequence.
In a few words, three program parameters are needed to build a model:
- The name of the queried graph: the graph URI (Uniform Resource Identifier),
- the name of the subset of data if it exists: the provenance URI,
- the endpoint: the the URL of the server.
Get the description of graphs on a triplestore¶
Example of Pathway Commons¶
When all databases are grouped under the same graph URI, filtering on the provenance is mandatory.
$ biopax2cadbiom model --getGraphUris --triplestore http://rdf.pathwaycommons.org/sparql/ Graph URI: http://pathwaycommons.org Provenance URI: http://pathwaycommons.org/pc2/biogrid Name: BioGRID Comment: Source type: PSI_MI, BioGRID Release 3.4.149 (human and the viruses), 25-May-2017 --- Graph URI: http://pathwaycommons.org Provenance URI: http://pathwaycommons.org/pc2/kegg Name: KEGG Pathway Comment: Source http://www.cogsys.cs.uni-tuebingen.de/mitarb/draeger/BioPAX.zip type: BIOPAX, KEGG 07/2011 (only human, hsa* files), converted to BioPAX by BioModels (http://www.ebi.ac.uk/biomodels-main/) team --- Graph URI: http://pathwaycommons.org Provenance URI: http://pathwaycommons.org/pc2/pid Name: NCI Pathway Interaction Database: Pathway Comment: Source type: BIOPAX, NCI Curated Human Pathways from PID (final); 27-Jul-2015 --- Graph URI: http://pathwaycommons.org Provenance URI: http://pathwaycommons.org/pc2/panther Name: PANTHER Pathway Comment: Source ftp://ftp.pantherdb.org/pathway/3.4.1/BioPAX.tar.gz type: BIOPAX, PANTHER Pathways 3.4.1 on 04-Jul-2016 (auto-converted to human-only model) --- Graph URI: http://pathwaycommons.org Provenance URI: http://pathwaycommons.org/pc2/reactome Name: Reactome Comment: Source http://www.reactome.org/download/current/biopax.zip type: BIOPAX, Reactome v61 (only 'Homo_sapiens.owl') 23-Jun-2017 --- ...Below is the command to make a Cadbiom model from Pathway Commons, based on PID:
$ biopax2cadbiom model \ --graph_uris http://pathwaycommons.org \ --provenance_uri http://pathwaycommons.org/pc2/pid \ --triplestore http://rdf.pathwaycommons.org/sparql/
- The parameter
--graph_urisprovides the URI of the graphs queried (and optionally of the BioPAX ontology if it is hosted separately). - It is thus necessary to filter the RDF triples according to their origin with the
the optional parameter:
--provenance_uri. By setting it, the program will filter entities, reactions, pathways thanks to theirdataSourceBioPAX attribute. - The URL of the endpoint is specified with
--triplestore.
Example of named graphs¶
When all databases are separated in different graphs, filtering on the provenance is NOT mandatory.
$ biopax2cadbiom model --getGraphUris --triplestore http://127.0.0.1:8890/sparql/ Graph URI: http://reactome.org/cavia Name: None Comment: http://www.reactome.org --- Graph URI: http://reactome.org/cricetulus Name: None Comment: http://www.reactome.org --- Graph URI: http://reactome.org/crithidia Name: None Comment: http://www.reactome.org --- Graph URI: http://reactome.org/escherichia Name: None Comment: http://www.reactome.org --- Graph URI: http://reactome.org/homarus Name: None Comment: http://www.reactome.org --- Graph URI: http://reactome.org/mycobacterium Name: None Comment: http://www.reactome.org --- Graph URI: http://reactome.org/triticum Name: None Comment: http://www.reactome.org --- Graph URI: http://reactome.org/vigna Name: None Comment: http://www.reactome.org --- Graph URI: http://www.irisa.fr/dyliss/data/metacyc20 Name: None Comment: None --- Graph URI: http://www.pathwaycommons.org/v9/pid Name: NCI Pathway Interaction Database: Pathway Comment: Source type: BIOPAX, NCI Curated Human Pathways from PID (final); 27-Jul-2015 --- Graph URI: http://biopax.org/lvl3 Name: None Comment: None --- Graph URI: http://virtualcases.org/1 Name: None Comment: None --- ...Below is the command to make a model from a local endpoint, hosting the PID graph:
$ biopax2cadbiom model \ --graph_uris http://biopax.org/lvl3 http://www.pathwaycommons.org/v9/pid \ --triplestore http://127.0.0.1:8890/sparql/
- The parameter
--graph_urisprovides the URI of the graphs queried (and optionally of the BioPAX ontology if it is hosted separately). - The URL of the endpoint is specified with
--triplestore.
Generated files¶
The conversion process will generate the model file (with a .bcx extension)
in the following folder: ./output.
Unless the --no_scc_fix option is specified, two models are designed.
The first is a default model corresponding to a strict conversion of the BioPAX content
into a formalism based on guarded transitions. The second has specific alterations
intended to optimize the operation of Cadbiom and its solver.
Strongly Connected Components¶
A Cadbiom model is considered as a directed graph, and is interpreted as such when it is loaded.
A directed graph is strongly connected if there is a path between all pairs of vertices in each direction. A strongly connected component (SCC) of a directed graph is a maximal strongly connected subgraph (any additonal edge breaks this property). For example, there are 2 SCCs in the following graph (we remove SCCs of 1 element to keep only “cycles”):
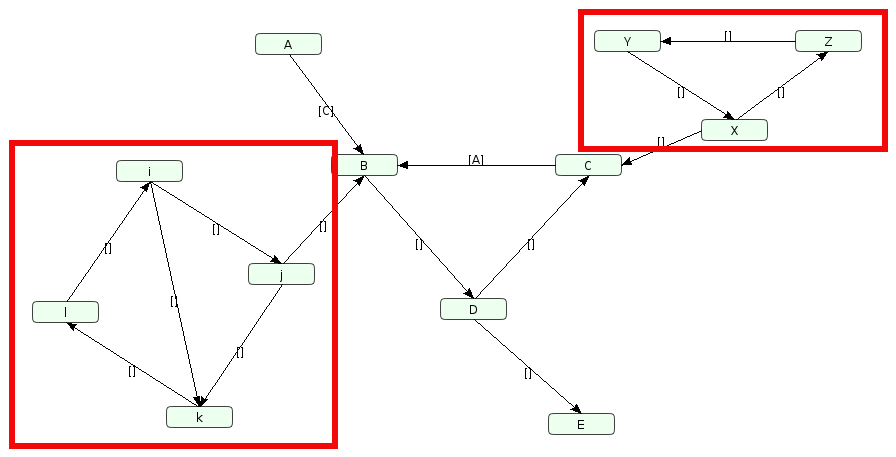
Two Strongly Connected Components on Cadbiom GUI: (i, j, k, l) and (X, Y, Z).
As it stands, there is no path from A to E without going through one of the two infinite loops (i, j, k, l) or (X, Y, Z). In order to unblock the path, we are forced to choose one node from each of the SCCs and convert it to a “start node”. The choice is systematically on the first node in the lexicographic order.
The --no_scc_fix option allows the creation of a new graph including these modifications.
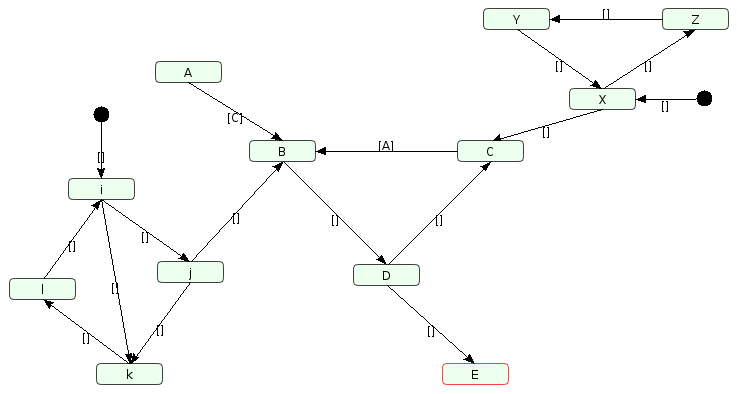
Removal of SCCs on Cadbiom GUI with the addition of two start nodes: i and X.
Note
Nodes selected as “start nodes” are considered as boundaries/frontier places.
Additional files¶
During the process 3 additional files are created in order to keep track of the changes and interpretations made to BioPAX data:
sort_dumped.txt: The sorted list of entities in the queried graph database. This file can also contain duplicate entities specially created for the purposes of data translation (see virtualcase11)sort_grouped.txt: Groups of entities with similar characteristics.sort_grouped_after_merge.txt: The final groups of entities, where we can see the definitive URI choosen for each group.