Examples¶
Examples of conversions from BioPAX graphs towards Cadbiom models¶
BioPAX (owl files) and Cadbiom models (bcx files) are hosted on our cloud: CeSGO
Reactome - Homarus_americanus¶
Command:
biopax2cadbiom model --graph_uris http://biopax.org/lvl3 http://reactome.org/homarus
Download link: homarus.bcx
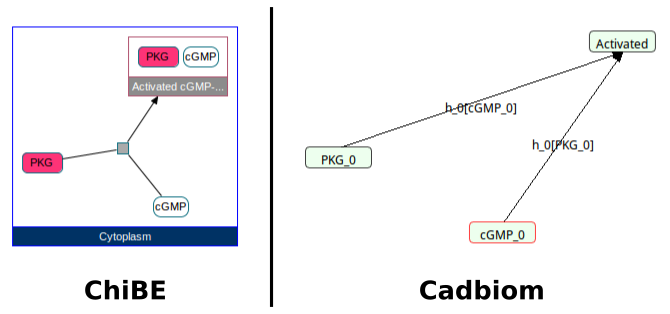
Reactome - Crithidia_fasciculata¶
Challenge: 1 BiochemicalReaction, 1 reagent, 2 products and 1 activator: Asat.
The activator must be in the conditions of both transitions.
Command:
biopax2cadbiom model --graph_uris http://biopax.org/lvl3 http://reactome.org/crithidia
Download link: crithidia.bcx
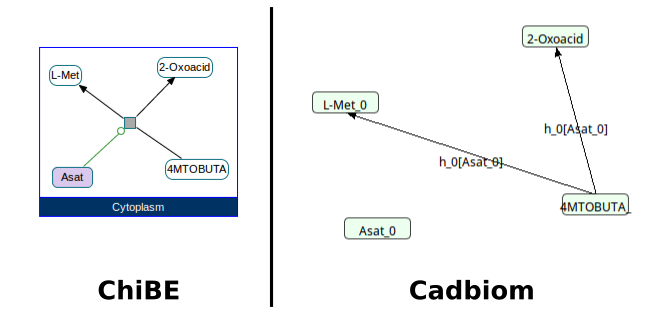
Reactome - Vigna_radiata_var._radiata¶
Challenge: Similar to Crithidia_fasciculata but with multiple products.
Command:
biopax2cadbiom model --graph_uris http://biopax.org/lvl3 http://reactome.org/vigna
Download link: vigna.bcx
Reactome - Triticum_aestivum¶
Command:
biopax2cadbiom model --graph_uris http://biopax.org/lvl3 http://reactome.org/triticum
Download link: triticum.bcx
Reactome - Cavia_porcellus¶
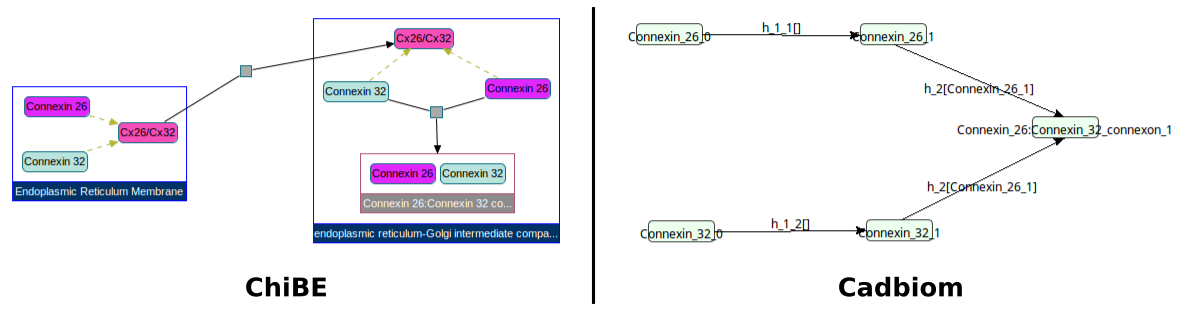
Challenge: Cx26/Cx32 is a class with 2 members: Connexin 26 and Connexin 32.
Each of these members belongs to 2 compartments: endoplasmic_reticulum_membrane and endoplasmic_reticulum_Golgi_intermediate_compartment.
The class itself has 1 version for both compartments (which may not be the case as we will see later,
the compartments are unequally specified in classes and members).
The goal is to remove the notion of class dedicated to visualization, and to connect
the versions of connexin between them. We must not have a direct transition from
Connexin26 to Connexin32 as this would defeat the underlying information.
In BioPAX data we have the following entities:
Protein1:
Protein2 Connexin_26_endoplasmic_reticulum_membrane
Protein3 Connexin_32_endoplasmic_reticulum_membrane
Protein4:
Protein5 Connexin_26_endoplasmic_reticulum_Golgi_intermediate_compartment
Protein6 Connexin_32_endoplasmic_reticulum_Golgi_intermediate_compartment
Complex1 Connexin_26_Connexin_32_connexon_endoplasmic_reticulum_Golgi_intermediate_compartment
Thus, the following transitions are not allowed:
Protein3 -> Protein5
Protein2 -> Protein6
Command:
biopax2cadbiom model --full_graph --graph_uris http://biopax.org/lvl3 http://reactome.org/cavia
Download link: cavia.bcx
Virtual Case 1¶
Short explanation about complex conditions in the formalism of guarded transitions:
Indeed, clock expressions uses default and when operators.
Operands of default operator must be of clock type.
The left operand of when operator must be of clock type while
the right one (the condition) must be a boolean expression similar
to the ones allowed in conditions.
Examples:
clock default
clock when (boolean expr)
default can operator merges 2 events:
h1 default h2
Thus, h1 when (boolean expr) defines a new event and h1 is triggered when the (boolean expr) is True.
Otherwise, the event is absent.
—
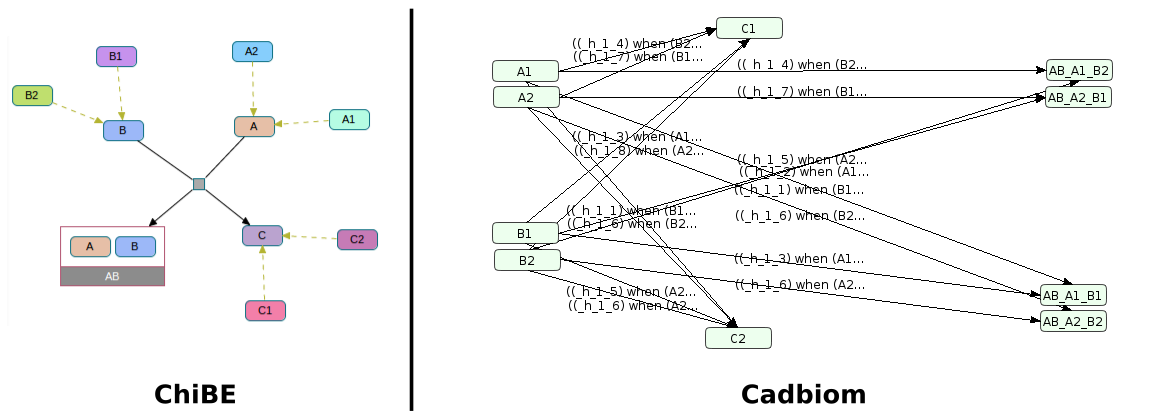
Challenge:
- 1 class
BcontainingB1andB2 - 1 class
AcontainingA1andA2 - 1 class
CcontainingC1andC2 - 1 reaction between classes:
A+B=> complexAB+C
Develop classes A and B and reconnect their child entities to products.
There is also a generation of the Cartesian product of the 4 transitions (A1B1, A1B2, A2B1, A2B2)
with C1 and C2.
PS: Without the --full_graph option, the module does not decompile classes
because their elements are not reused elsewhere in the model.
Command:
biopax2cadbiom model --full_graph --graph_uris http://biopax.org/lvl3 http://virtualcases.org/1
Download link: virtualCase1.bcx
Virtual Case 2¶
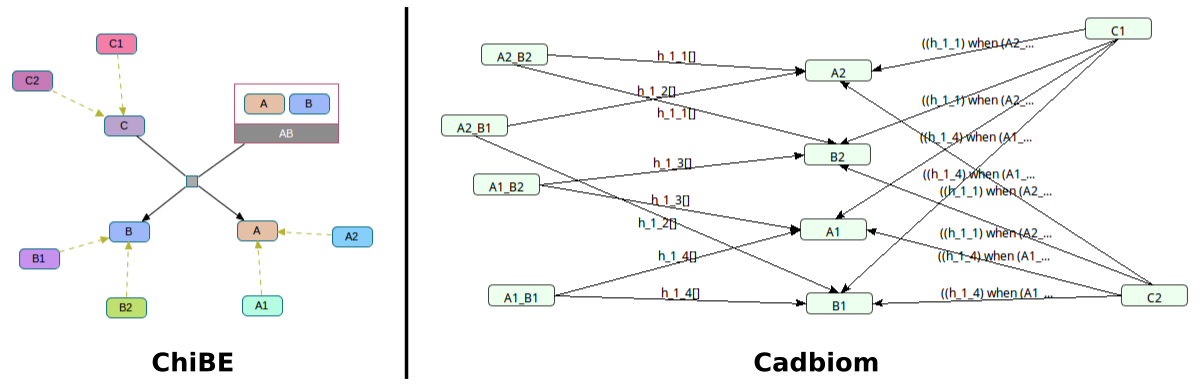
Challenge: Decompilation of a complex with used elements.
1 class C + 1 complex AB of 2 classes A and B => 2 classes A and B
with disappearance of the class C.
C1+A2_B2=>A2+B2C2+A1_B1=>A1+B1- etc.
Command:
biopax2cadbiom model --full_graph --graph_uris http://biopax.org/lvl3 http://virtualcases.org/2
Download link: virtualCase2.bcx
Virtual Case 1-2b¶
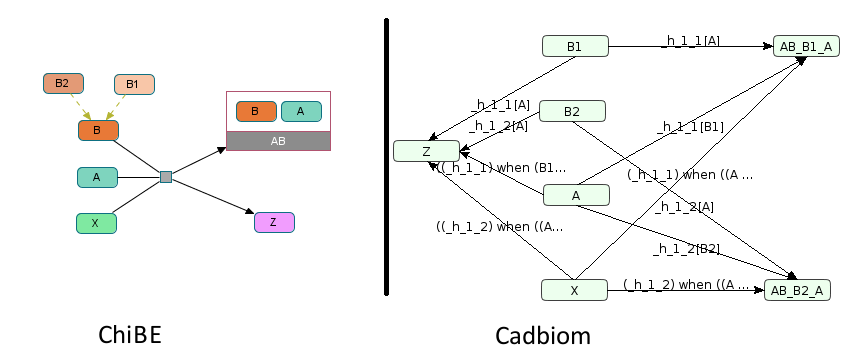
Challenge: Connect entities without antecedent (the product Z),
and without successor (the reactant X). These entities are not classes
as in the cases virtualCase1 and virtualCase2.
The magic is done in the definition of events:
X -> Z: ((_h_1_2) when ((A and B2))) default ((_h_1_1) when ((A and B1)))
X -> AB_B1_A: (_h_1_1) when ((A and B1))
X -> AB_B2_A: (_h_1_2) when ((A and B2))
Command:
biopax2cadbiom model --full_graph --graph_uris http://biopax.org/lvl3 http://virtualcases.org/1-2b
Download link: virtualCase1-2b.bcx
Reactome - Escherichia_coli¶
Challenge: A class in a complex with 20 members. None of its members are reused elsewhere in the model. The class itself is lost in the complex obtained in product of the reaction.
With option --full_graph, we must have 20 reactions (obtained by duplication);
otherwise the class is preserved and only one reaction remains in the final model.
Command:
biopax2cadbiom model --full_graph --graph_uris http://biopax.org/lvl3 http://reactome.org/escherichia
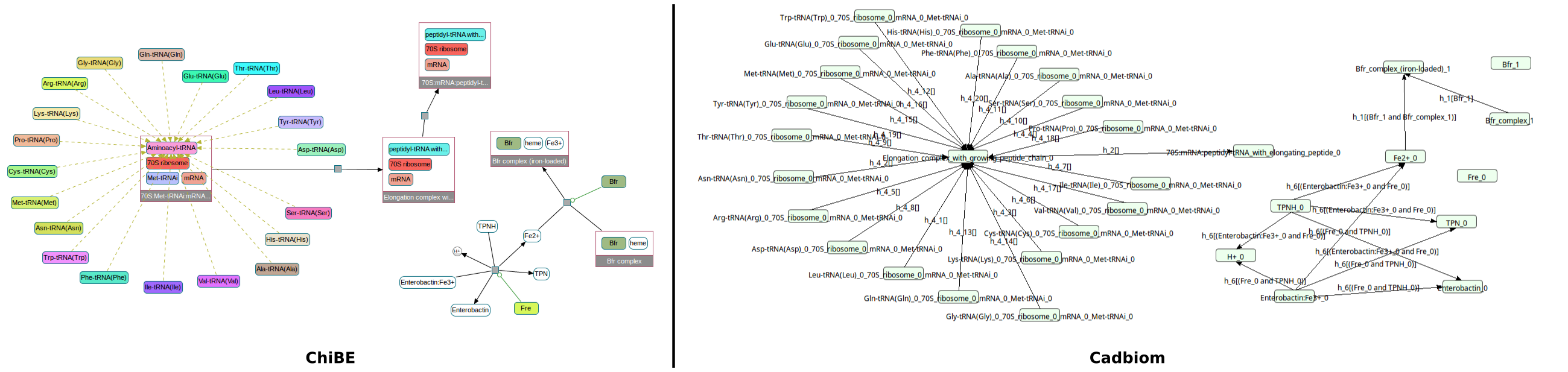
Download link: escherichia.bcx
Reactome - Cricetulus_griseus¶
Command:
biopax2cadbiom model --full_graph --graph_uris http://biopax.org/lvl3 http://reactome.org/cricetulus
Download link: cricetulus.bcx
Reactome - Mycobacterium_tuberculosis¶
Command:
biopax2cadbiom model --full_graph --graph_uris http://biopax.org/lvl3 http://reactome.org/mycobacterium
Download link: mycobacterium.bcx
Virtual Case 3¶
Challenge: Decompilation of a class with useless elements.
One class A of which only one element A1 is reused elsewhere as a modulator.
This element is considered as an entity. The others are ignored.
Command:
biopax2cadbiom model --graph_uris http://biopax.org/lvl3 http://virtualcases.org/3
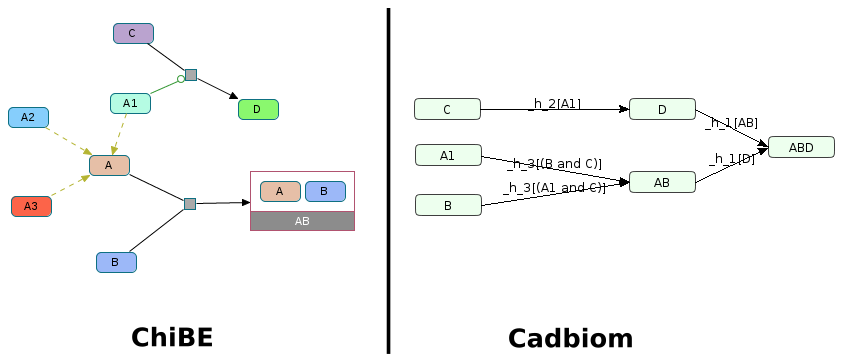
Download link: virtualCase3.bcx
Virtual Case 4¶
Challenge: 2 ComplexAssembly reactions using a class with 3 members. We should obtain 3 reactions for both of these reactions; thus 6 reactions.
Note: If --full_graph is not set we will have the following condition:
(A | B) & ~(C | D)
Command:
biopax2cadbiom model --full_graph --graph_uris http://biopax.org/lvl3 http://virtualcases.org/4
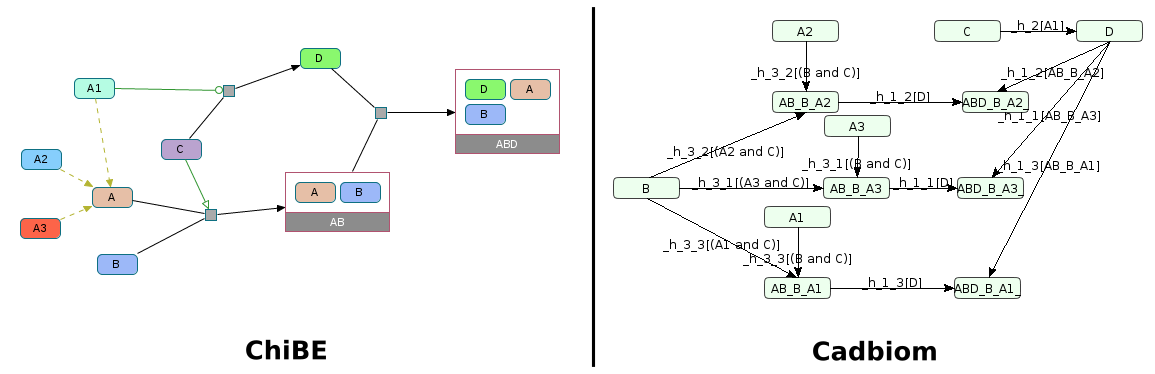
Download link: virtualCase4.bcx
Virtual Case 5¶
Challenge: Combination of modulators and major rule of interpretation of their combinations.
- 2 classes
AandCacting on a reaction:A(activator),C(inhibitor) - 2 other modulators on the same reaction: B (activator), D (inhibitor)
We want at least 1 activator AND NO inhibitor for the reaction to take place.
Command:
biopax2cadbiom model --full_graph --graph_uris http://biopax.org/lvl3 http://virtualcases.org/5
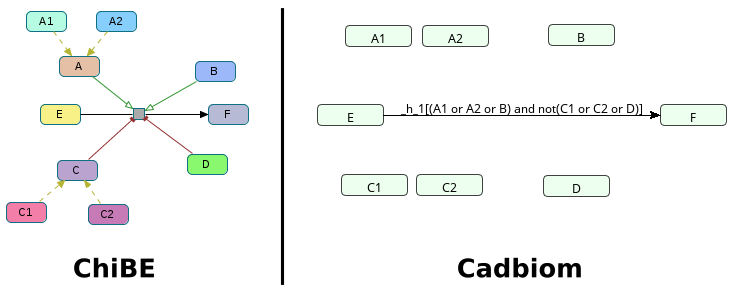
Download link: virtualCase5.bcx
Virtual Case 6¶
Challenge: A class A with 2 members A1 and A2 that are not
reused elsewhere. A is a component of the complex AB that
regulates the reaction C -> D. Without the flag --full_graph, A1
and A2 will not be found in the model.
The conditions of the transitions are calculated accordingly:
B -[A]> AB
C -[AB]> D
or
B -[A1]> A1B
B -[A2]> A2B
C -[A1B or A2B]> D
Command:
biopax2cadbiom model --graph_uris http://biopax.org/lvl3 http://virtualcases.org/6
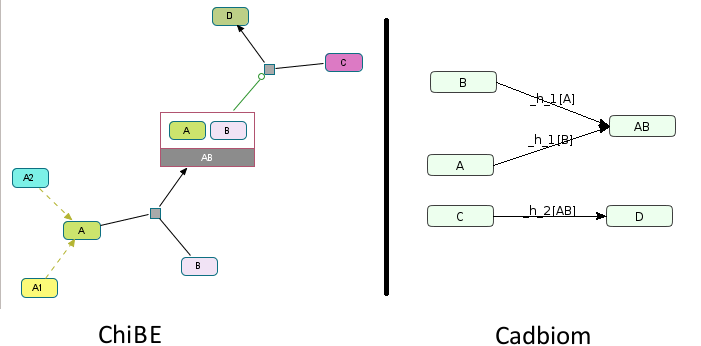
Download link: virtualCase6.bcx
Command:
biopax2cadbiom model --full_graph --graph_uris http://biopax.org/lvl3 http://virtualcases.org/6
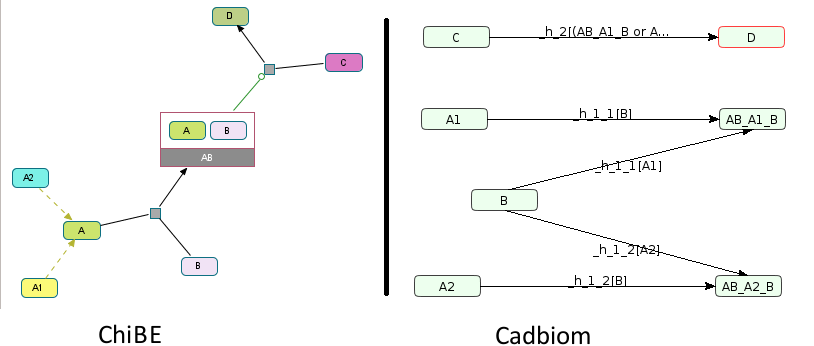
Download link: virtualCase6a.bcx
Virtual Case 6b¶
Challenge: Same as virtualCase6 but A is a simple protein instead a class.
Command:
biopax2cadbiom model --graph_uris http://biopax.org/lvl3 http://virtualcases.org/6b
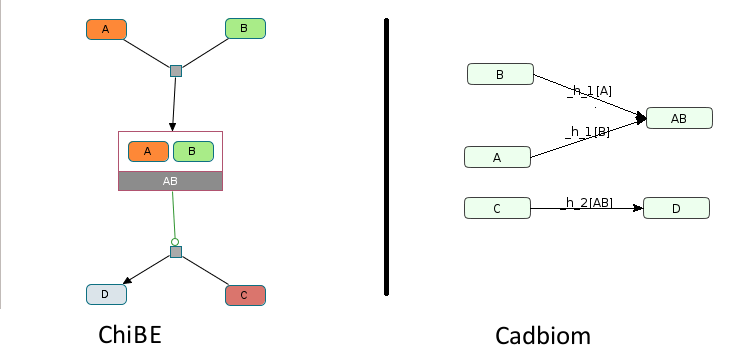
Download link: virtualCase6b.bcx
Virtual Case 7¶
Challenge: A class A with 3 members A1, A2 and A3;
A1 and A3 are reused elsewhere. A1 is a control of the
reaction B -> C. A3 participates to the reaction A3 -> D and
A participates to the reaction A -> E. Without
–full_graph, A2 must not be present in the model.
PS: Until now in test cases, multiple members of classes were not reused in the model.
Command:
biopax2cadbiom model --graph_uris http://biopax.org/lvl3 http://virtualcases.org/7
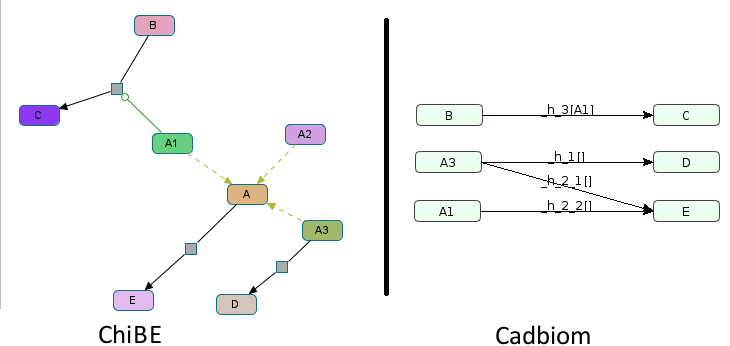
Download link: virtualCase7.bcx
Virtual Case 8¶
Challenge: Naming of entities according to their BioPAX ModificationFeatures.
ModificationFeatures are covalent bindings between entities that are not proteins in theory;
in practice, they can symbolize the addition of an inorganic phosphorus, an acetyl group or a protein of ubiquitin.
More generally, these are post-translational modifications.
The ModificationFeatures help us to uniquely name cadbiom entities that can have the same standard name.
A can have 3 states: phosphorylated once, phosphorylated twice,
phosphorylated twice and ubiquitinated and acetylated simultaneously.
Command:
biopax2cadbiom model --graph_uris http://biopax.org/lvl3 http://virtualcases.org/8
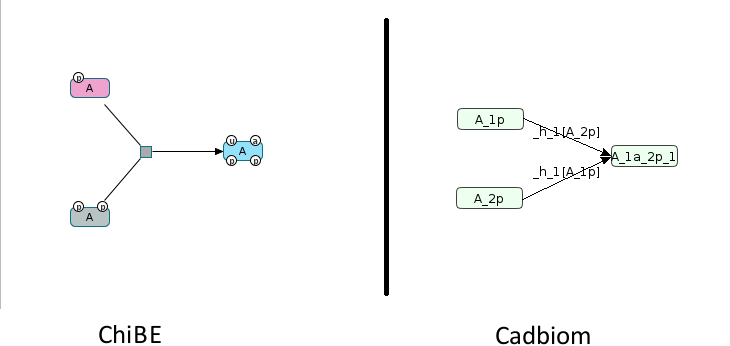
Download link: virtualCase8.bcx
Virtual Case 9¶
First challenge: The naming of entities that inherit classes according to their BioPAX ModificationFeatures attribute. Classes can carry modifications and locations that must be transferred to child entities.
Here, Class_A-B has one modification: residue modification active; during the suppression of the genericity A and B entities must inherit of it.
Command:
biopax2cadbiom model --full_graph --graph_uris http://biopax.org/lvl3 http://virtualcases.org/9b
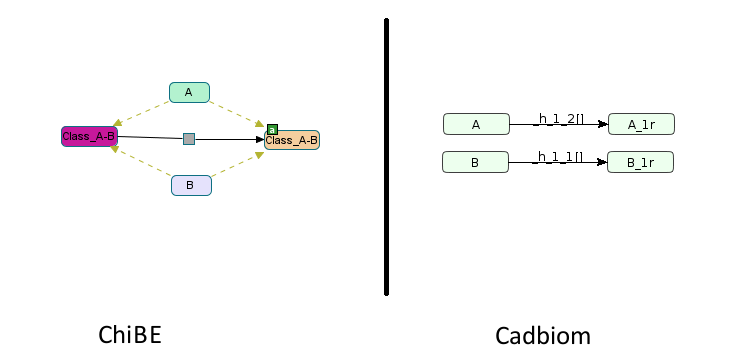
Download link: virtualCase9b.bcx
Second challenge: The identification of similar entities on both sides of the reaction after the suppression of the genericity (already seen in the case of test virtualcase1).
A and B have specific entityReferences in the previous example.
Because the names are different on both sides, without this BioPAX attribute
the program can’t know that A is only linked to A_1r , it
is thus obligatory to make the cartesian product between all entities.
The following command illustrates the cartesian product made when there is no entityReferences in the entities.
Command:
biopax2cadbiom model --full_graph --graph_uris http://biopax.org/lvl3 http://virtualcases.org/9
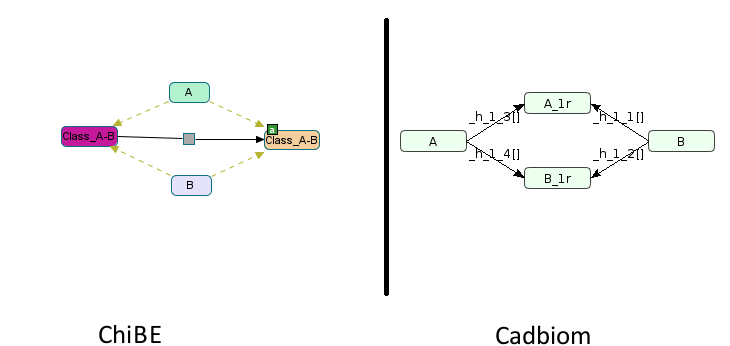
Download link: virtualCase9.bcx
Virtual Case 10¶
Challenge: Handle self-generating class with 0, 1 or more members reused elsewhere. Reactions can be converted into transitions from BioPAX but should not be loaded into the model. Moreover, since these are the same molecules on both sides, we do not want a cartesian product between the members of the class.
- a: 0 member,
Ais a self-generating class without reused member.
Command:
biopax2cadbiom model --graph_uris http://biopax.org/lvl3 http://virtualcases.org/10
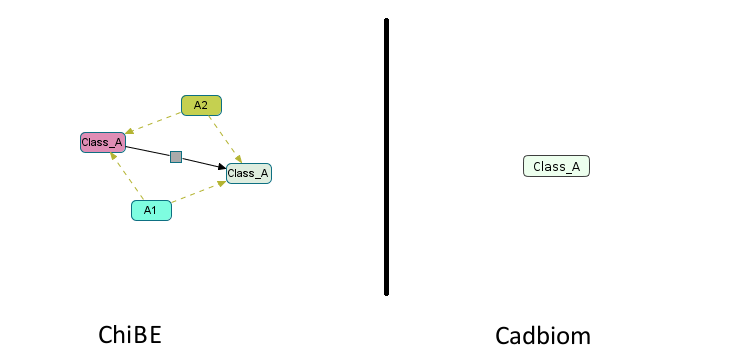
Download link: virtualCase10.bcx
Note: A warning appears when loading the model:
WARNING: Reflexive transition: Class_A -> Class_A - This transition will not be taken into account.
- b: 1 member,
Ais a self-generating class with one reused member.
Command:
biopax2cadbiom model --graph_uris http://biopax.org/lvl3 http://virtualcases.org/10b

Download link: virtualCase10b.bcx
Note: A warning appears when loading the model:
WARNING: Reflexive transition: A1 -> A1 - This transition will not be taken into account.
- c: 2 members,
Ais a self-generating class with all its members reused.
Command:
biopax2cadbiom model --full_graph --graph_uris http://biopax.org/lvl3 http://virtualcases.org/10
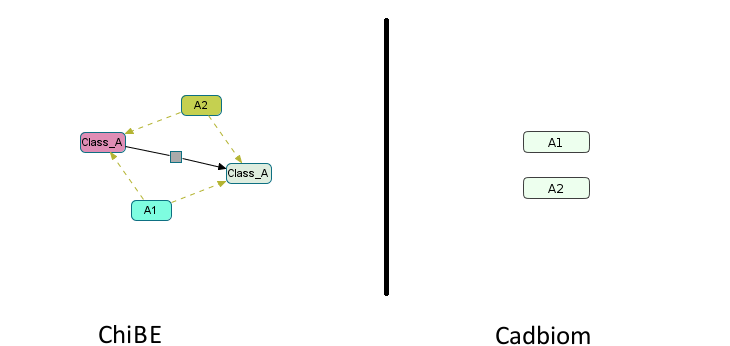
Download link: virtualCase10c.bcx
Note: Two warnings appear when loading the model:
WARNING: Reflexive transition: A1 -> A1 - This transition will not be taken into account.
WARNING: Reflexive transition: A2 -> A2 - This transition will not be taken into account.
Virtual Case 11¶
Challenge: Identify and merge together the duplicated entities in the model. Please note that the duplicates can come from the BioPAX database, as well as from the process of transferring post-translational modifications of classes to their daughter entities in transfer_class_attributes_on_child_entities function
In this example there are 2 similar complexes with different URIs: #Complex_Aa and #Complex_Abisa.
They are called similar because they share the same following attributes:
- entityType:
Complex- entityRef:
None- name:
A(dimer)- components_uris:
#Protein_A- location_uri:
#CellularLocationVocabulary_nucleus- modificationFeatures:
#ModificationFeature_activation
There is an inactivation reaction of the complex #Complex_Aa to the complex #Complex_Ai,
and a reaction of formation of the complex named A(dimer)-B based on a protein B and the complex #Complex_Abisa.
Merging entities brings together the complex #Complex_Aa and #Complex_Abisa as a single entity.
Note: As we can see on the following illustration, Chibe does not represent homomeric complexes the same way that heteromeric complexes, and omits the representation of the activation or inactivation information of them.
Note: During the conversion process 3 additional files are created in order to keep track of the changes made:
sort_dumped.txt: The sorted list of entities in the queried graph database. This file can also contain duplicate entities specially created for the purposes of data translation.sort_grouped.txt: Groups of entities with similar characteristicssort_grouped_after_merge.txt: The final groups of entities, where we can see the definitive URI choosen for each group.
Command:
biopax2cadbiom model --graph_uris http://biopax.org/lvl3 http://virtualcases.org/11
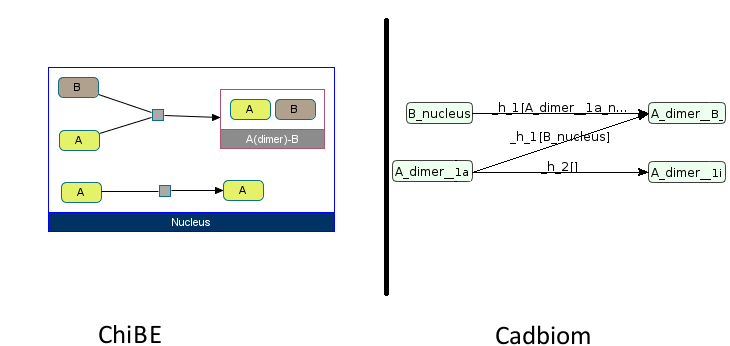
Download link: virtualCase11.bcx
Virtual Case 14¶
Challenge: The decompilation of a self-generating class must not generate erroneous transitions.
A: A class with 2 members A1, A2.
The BiochemicalReaction transforms the class A, giving a class Aa (with a modification feature) and vice versa.
Implicit presence of the entities A2_1a and A1 in the model;
Explicit presence of A1_1a and A2`` in the model.
When transferring the modifications of each class to its members, if we delete the reactions in which the members participate:
Decompiling
AgivesA1andA2, sinceA2is the only reused entity of the class, it takes the place ofA. DecompilationAagivesA1aandA2a, sinceA1ais the only reused entity of the class, it takes the place ofAa. Thus in the final model,A = A2andAa = A1a, thereforeA2 <=> A1a. => false reactionOtherwise, we admit that
A2ainherits the reaction ofAaduring its decompilation:However the presence of
A1awill not be impacted because it already participates in 1 reaction (B => A1a). In this case, sinceA1is not used in the model, the reactionA1 <=> A1awill not exist for the sake of simplification.
Full decompilation command:
biopax2cadbiom model --full_graph --graph_uris http://biopax.org/lvl3 http://virtualcases.org/14
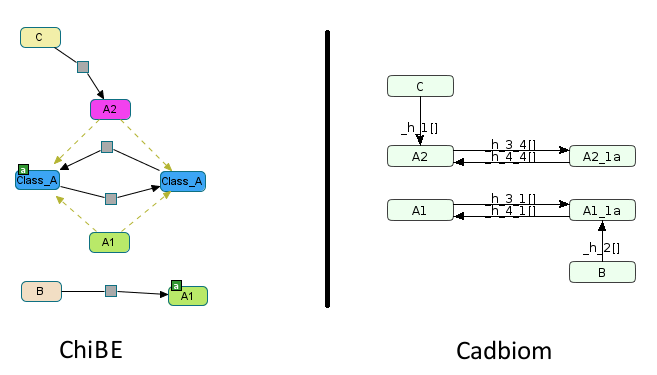
Download link: virtualCase14.bcx
Automatic decompilation command:
biopax2cadbiom model --graph_uris http://biopax.org/lvl3 http://virtualcases.org/14b
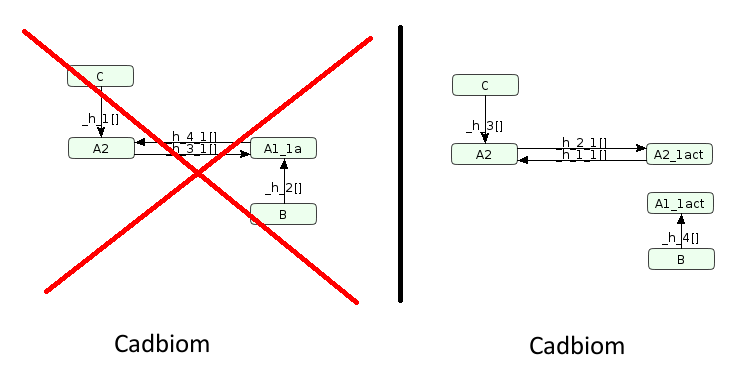
Download link: virtualCase14b.bcx
We can handle more complex cases:
We test the behavior of an entity merge on a similar model except for the addition of a copy of the entity
A2a (Protein_A2bis) that doesn’t participate in any reaction.
Reminder: A2a does not exist in the original model, it is created from scratch.
Duplication of A2 once it has inherited the modifications from its parent class Aa
leads to a duplicated entity which also inherit of the reaction A <=> Aa.
When grouping the entities, A2a and Protein_A2bis will be merged.
The attribution of a reaction to this group, coming only from the duplicated entity
is not an argument for its removal because this will reproduce the problem showed in virtualCase14
(false transition A2 <=> A1a).
Automatic decompilation with merge of entities command:
biopax2cadbiom model --graph_uris http://biopax.org/lvl3 http://virtualcases.org/14c
Note: The final model is the same as with the previous command.
Virtual Case 17¶
Challenge: Support of complexes with entities which are also in their components that are classes. See Virtual Case 19 for the support of complexes with similar classes in their nested components.
A: A class with 2 members A1, A2,
AA1: Complex with the following components:
Aclass with 2 membersA1,A2,A1simple complex.
So A1 is also in A. During its suppression, we will obtain the following complexes:
A1A2
A1A1
Note: The complex A1A2 might not make sense.
This is why when we find the same class several times through the components of a complex,
we avoid making the Cartesian product between the members of this class. (See Virtual Case 19 )
In other words: We process multiple occurrences of classes but do nothing when members of these classes are found alone (out of their classes) in the components.
Command:
biopax2cadbiom model --full_graph --graph_uris http://biopax.org/lvl3 http://virtualcases.org/17
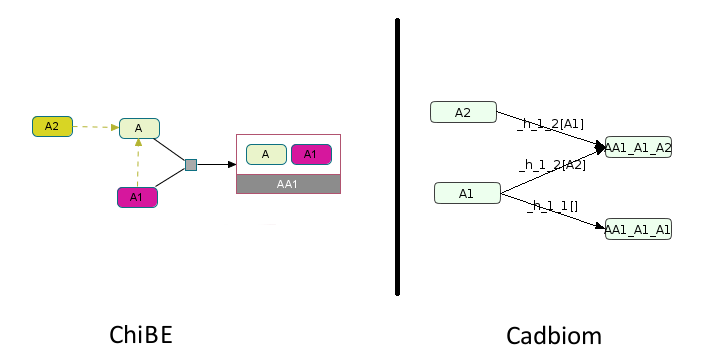
Download link: virtualCase17.bcx
Virtual Case 18¶
Challenge: Handle nested complexes; some of which may involve classes.
A_top: Complex with the following components (1 simple entity and 2 complexes):
A_bottom, complex with a componentAwhich is a class with the membersA1, andA2,A_bottom, complex with a componentAwhich is a class with the membersB1, andB2,Y, simple entity.
The BiochemicalReaction must be duplicated into 4 versions;
each giving a version of the complex A_top, made up of the entity Y
and one of the 2 members of the class of each sub-complex.
Command:
biopax2cadbiom model --full_graph --graph_uris http://biopax.org/lvl3 http://virtualcases.org/18
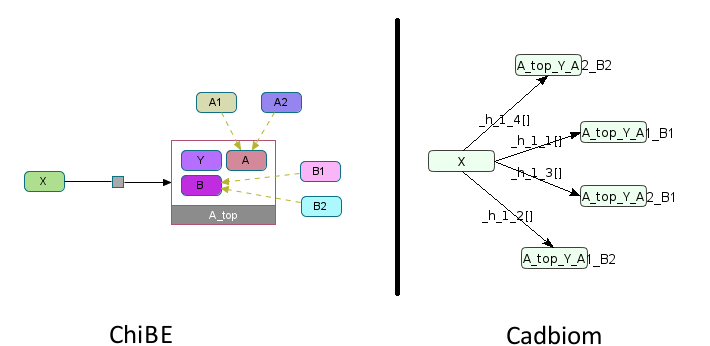
Download link: virtualCase18.bcx
Virtual Case 19¶
First challenge: Handle hybrid classes/complexes entities.
A_top: Complex with the following components:
Y, simple entity,B_bottomempty complex similar to a simple entity,A_bottom, hybrid class/complex.
A_bottom: Class with the following, 2 members and 1 component:
AA_bottom, simple Complex, reused in a reaction,AAA_bottom, simple Complex,X, simple entity as component.
The presence of a component in A_bottom that is supposed to be a class is problematic.
So we create a simple complex entity A_bottom_v2 (not_class) with the component X;
while we remove X from A_bottom which becomes a full class without component.
Generated complexes:
A_top_Y_AAA_bottom_B_bottom
A_top_Y_AA_bottom_B_bottom
A_top_Y_A_bottom_v2_B_bottom
Note: In Chibe, A_top is not represented as having the class A_bottom.
Only the X entity from A_bottom is displayed in A_top.
In other words, the simple complex nature overrides the generic nature of A_bottom
when it comes to representing it in Chibe.
—
Second challenge: Handle complexes with similar classes in their nested components. See Virtual Case 17 for the support of complexes with entities which are also in their components that are classes.
C_top: Complex with the following components:
Bclass with the membersB1,B2,C_bottomsimple complex.
C_bottom: Complex with a component B, which is a class with the members B1, and B2.
The flats components of C_top therefore contain twice the class B which will be replaced by its members.
We must not make the Cartesian product here: among the 4 combinations we only keep:
B1B1
B2B2
Indeed, the following combinations might not make sense:
B1B2
B2B1
See replace_and_build function where all class instances are replaced simultaneously by the same member.
Command:
biopax2cadbiom model --full_graph --graph_uris http://biopax.org/lvl3 http://virtualcases.org/19
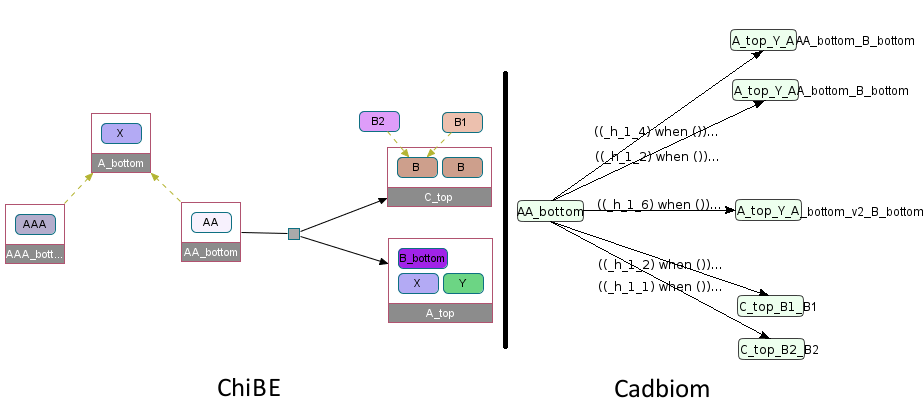
Download link: virtualCase19.bcx
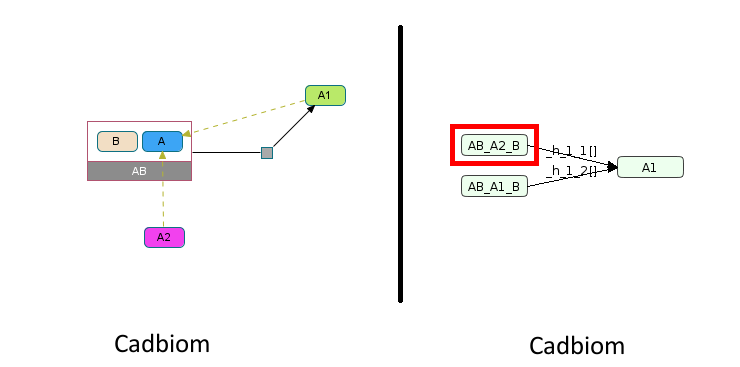
Virtual Case 20¶
First challenge: Support for a partially used classes.
A: Complex with the following components:
A1,A2
A reaction involving a class A as a reagent and only 1 member of that class as a product A1.
Duplication of the reaction for each member (A1 and A2) induces the creation of an erroneous reaction:
A1B -> A2.
Indeed, biopax2cadbiom.reactions.get_banned_reagents_products_couples()
is only interested in the classes present on both sides of the reaction.
The couples of banned reactants are generated according to the content of the found classes.
If there is no class, there are no banned pairs.
This case can happen, but it is more a formalization/representation error in the database than a problem to address.
Command:
biopax2cadbiom model --full_graph --graph_uris http://biopax.org/lvl3 http://virtualcases.org/20
Download link: virtualCase20.bcx
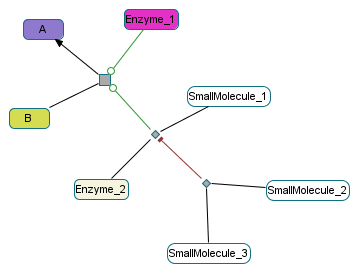
Control 1¶
Challenge: Handle multiple regulations for 1 reaction (see Virtual Case 5); Handle multiple controllers per control; Handle cascade of controls.
SmallMolecule_1 is a cofactor of Enzyme_2; both are controllers of the same Control object which activates a BiochemicalReaction
A -> B.
SmallMolecule_2 and SmallMolecule_3 are the controllers of another Control object which inhibits the previous Control.
Expected condition for the transition A -> B:
Enzyme_1 or (Enzyme_2 and SmallMolecule_1 and not (SmallMolecule_2 and SmallMolecule_3))
Command:
biopax2cadbiom model --graph_uris http://biopax.org/lvl3 http://virtualcases.org/test_control_1
Download link: test_control1.bcx
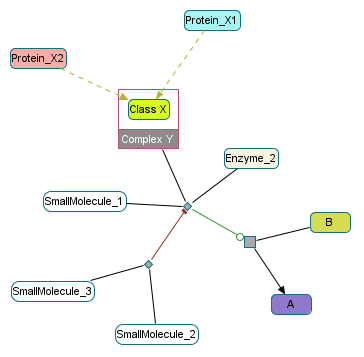
Control 2¶
Challenge: Similar to Control 1 except that the control of the BiochemicalReaction accepts one more controller: a Complex composed by a class.
Y: Homodimeric Complex with the following component:
X, A class with 2 membersX1,X2
Expected condition for the transition A -> B:
Enzyme_2 and SmallMolecule_1 and (Complex_Y_Protein_X1 or Complex_Y_Protein_X2) and not (SmallMolecule_2 and SmallMolecule_3)
Command:
biopax2cadbiom model --full_graph --graph_uris http://biopax.org/lvl3 http://virtualcases.org/test_control_2
Download link: test_control2.bcx