Troubleshooting¶
Verbose and debug output¶
Biopax2Cadbiom comes with a build feature that can outputs a lot of debug information when enabled.
Logs are stored in /tmp/biopax2cadbiom.log on GNU/Linux. By default the logging level outputs only
warnings and errors, but the verbosity can be increased with a similar command:
$ biopax2cadbiom -vv debug ...
Conversion issues¶
Processing BioPAX files produced with the Cytoscape BiNOM Plugin¶
In order to analye the ACSN file “ACSN 2.0 Master.xml”, we used the BioPAX files produced by the BiNoM cytoscape plugin. As the original file was not compatible with the BiNoM plugin, we processed and analyzed all individual maps provided by the ACSN website (adaptive immune, angiogenesis_master, caf cell, cellcycle dnarepair, dendritic cell, innate immune, invasion motility, macrophages mdsc cells, natural killer cell, rcd, survival, telomere maintenance). The BioPAX files produced by the BiNoM cytoscape plugin are characterized by the presence in each of the BioPAX objects of the whole hierarchy of the parent classes in the ontology. Each object is mostly described according to the most generic type possible (Entity, Conversion, Interaction, Control). The most precise type of the object is actually described in the object properties via rdf:type. The main issue generated by this format is that when we query objects with the rdfs:subClassOf* property (rdfs:subClassOf is an instance of rdf:Property), objects appear as many times as they have parent classes. In addition, since the BioPAX superclass is “Entity” reaction can be considered as entities.
Example of Protein:
<bp:PhysicalEntity rdf:about="http://www.biopax.org/release/biopax-level3.owl#PRKCZ_at_Cytosol">
<bp:name rdf:datatype="http://www.w3.org/2001/XMLSchema#string">PRKCZ@Cytosol</bp:name>
<rdf:type rdf:resource="http://www.biopax.org/release/biopax-level3.owl#Entity"/>
<rdf:type rdf:resource="http://www.biopax.org/release/biopax-level3.owl#Protein"/>
</bp:PhysicalEntity>
Example of Catalysis:
<bp:Control rdf:about="http://www.biopax.org/release/biopax-level3.owl#emtc_emtc_re28_c_emtc_emtc_s3983">
<bp:controlType rdf:datatype="http://www.w3.org/2001/XMLSchema#string">INHIBITION</bp:controlType>
<bp:controller rdf:resource="http://www.biopax.org/release/biopax-level3.owl#CTBP1_2__HDAC1_HDAC3_SNAI2_at_Nucleus"/>
<bp:controlled rdf:resource="http://www.biopax.org/release/biopax-level3.owl#emtc_emtc_re28"/>
<rdf:type rdf:resource="http://www.biopax.org/release/biopax-level3.owl#Interaction"/>
<rdf:type rdf:resource="http://www.biopax.org/release/biopax-level3.owl#Entity"/>
<rdf:type rdf:resource="http://www.biopax.org/release/biopax-level3.owl#Catalysis"/>
</bp:Control>
Another issue is related to redundancies on the type of string properties, such as in the following example:
<bp:controlType rdf:datatype="http://www.w3.org/2001/XMLSchema#string">ACTIVATION^^http://www.w3.org/2001/XMLSchema#string</bp:controlType>
To process the BioPAX files generated by BiNOM, which defines the entire hierarchy of parent classes for each BioPAX object, we ensured that objects have the most accurate interactionType attribute as possible. In practice Virtuoso returns first the rdf: type most accurate property, then the parent classes (Ex: BiochemicalReaction then Conversion in the case of an object that would include these 2 properties).
The complete procedure includes the following steps. (1) Production of remastered maps in which each species or reaction identifier has been prefixed with the name of the map. (cf process_celldesigner_maps.py) (2) Via Binom, export of owl files (3) Cleaning of redundant string types (~4000) via sed (Cf import_tests_to_database.sh)
sed -i -e 's/\^\^http:\/\/www.w3.org\/2001\/XMLSchema#string//g' *.owl
- Importing owl files
- Optional removal of redundant triplets for all parent classes in ontology:
DELETE { GRAPH <http://acsn.curie.fr/global> { ?reaction rdf:type ?reactionType }}
WHERE {
?reaction rdf:type ?reactionType .
?reactionType rdfs:subClassOf* biopax3:Interaction .
?reaction rdf:type ?reactionType2 .
?reactionType2 rdfs:subClassOf ?reactionType .
}
- Deletion of redundant types also for entities:
DELETE { GRAPH <http://acsn.curie.fr/global> { ?entity rdf:type ?entityType }}
WHERE {
?entity rdf:type ?entityType .
?entityType rdfs:subClassOf* biopax3:Entity .
?entity rdf:type ?entityType2 .
?entityType2 rdfs:subClassOf ?entityType .
}
Warnings and permanent nodes¶
During the conversion, some warnings with the following format can be generated:
WARNING: Reflexive transition: <ENTITY> -> <ENTITY> - This transition will not be taken into account.
Please review your model or use a PermanentNode instead.
This means that a transition reflects an autogeneration of an entity in the translated network. This is not usually supposed to happen and when the Cadbiom model is loaded, these transitions will not be interpreted.
During the development, these warnings help to solve and identify the unsupported cases of the BioPAX standard. However, in a release version, these are often inconsistencies in the BioPAX file. In the following you will find a number of real cases accompanied by explanations.
Inconsistencies in ModificationFeatures¶
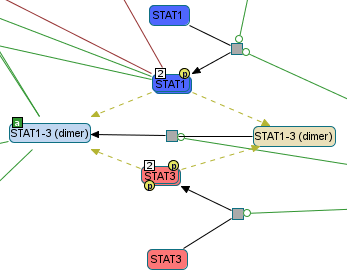
Screenshot of Chibe software; pathway IL27-mediated signaling events; file PathwayCommons9.pid.BIOPAX.owl available on http://pathwaycommons.org/.
STAT1-3 (dimer) that contains two dimeric complexes: STAT1, STAT3
(ids: Complex_4db5f804f0956c670c3408800208ecd7, Complex_177aaa44ccf4e366dd236d4775e2f28c).
However, each complexes already have a ModificationFeatures characterizing their activation.
Therefore when decompiling classes, and applying their own modifications to their child entities,
we obtain two auto-generating reactions for respectively STAT1__dimer__1a_v1 and STAT3__dimer___1a.
The problem is that dimers should not have posttranslational modifications here.Useless reaction 1¶
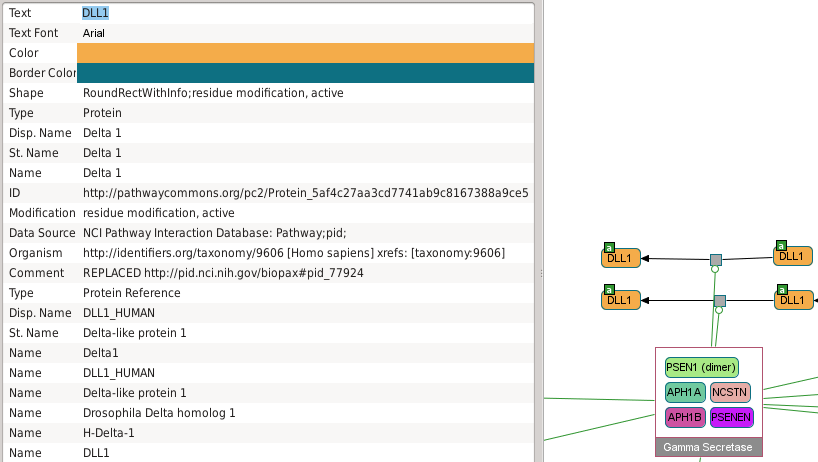
Screenshot of Chibe software; pathway Notch signaling pathway; file PathwayCommons9.pid.BIOPAX.owl available on http://pathwaycommons.org/.
Delta_1_1a -> Delta_1_1a and sDelta_1_1a -> sDelta_1_1a transitions are useless in the final model.
Note that Chibe does not distinguish sDelta_1_1a and Delta_1_1a and displays the name
of the gene instead (DLL1).Useless reaction 2¶

Screenshot of Chibe software; pathway FAS (CD95) signaling pathway; reaction BiochemicalReaction_e119b10c1af4f3e8892e2b3f4e70eb1f; file PathwayCommons9.pid.BIOPAX.owl available on http://pathwaycommons.org/.
FASLG_FAS__trimer__FADD_FADD_CASP8_CASP8_1a_integral_to_membrane_v2 -> FASLG_FAS__trimer__FADD_FADD_CASP8_CASP8_1a_integral_to_membrane_v2Useless reaction 3¶

Screenshot of Chibe software; pathway Regulation of nuclear SMAD2/3 signaling; reaction BiochemicalReaction_751f22441b4e03560c7a85f39efbac35; file PathwayCommons9.pid.BIOPAX.owl available on http://pathwaycommons.org/.
SMAD3_SMAD4_1a_nucleus -> SMAD3_SMAD4_1a_nucleusUseless reaction 4¶

Screenshot of Chibe software; pathway Role of Calcineurin-dependent NFAT signaling in lymphocytes; reaction BiochemicalReaction_a66d82dae0ef3db5c6a50da02b372567; file PathwayCommons9.pid.BIOPAX.owl available on http://pathwaycommons.org/.
NFAT1_1O_4O_1a_nucleus -> NFAT1_1O_4O_1a_nucleusUseless reaction 5¶
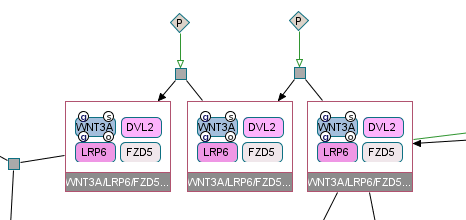
Screenshot of Chibe software; pathway Canonical Wnt signaling pathway; reaction BiochemicalReaction_881effb0cf3e3124280b7844f03669cc; file PathwayCommons9.pid.BIOPAX.owl available on http://pathwaycommons.org/.
WNT3A_LRP6_FZD5_DVL_1a_integral_to_membrane_v2 -> WNT3A_LRP6_FZD5_DVL_1a_integral_to_membrane_v2Useless reaction 6¶
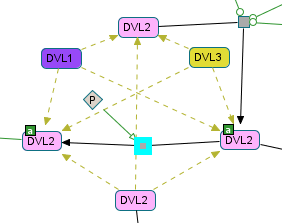
Screenshot of Chibe software; pathway Noncanonical Wnt signaling pathway; reaction BiochemicalReaction_23d1312ee8c92c4d5434cdc3f0985741; file PathwayCommons9.pid.BIOPAX.owl available on http://pathwaycommons.org/.
DVL2_1a is a self-generating class. One of its child entities is reused elsewhere in the model
(we see one of these reactions at the bottom of the screenshot: DVL2 -> ?).
A similar case is tested in virtualCase10c.Unsupported interactions¶
There may be errors related to the translation of some interactions defined in the BioPAX formalism.
Omitted reactions¶
Type of reaction is deliberately unsupported because it is too imprecise: <BIOPAX_URI>According to the BioPAX standard [1],
MolecularInteractionsare produced by high throughput systems that does not satisfy the level of detail (i.e. enough experimental evidence) required to model them withComplexAssemblyclass. These interactions will not be translated.
Screenshot of Chibe software; pathway N-Glycan biosynthesis; file PathwayCommons10.kegg.BIOPAX.owl available on http://pathwaycommons.org/.
We can see two MolecularInteractions between ALG3 and ALG9, and between ALG9 and ALG13.
Unexpected reactions¶
UNEXCEPTED REACTION: <BIOPAX_URI>During the development of Biopax2Cadbiom, we have never encountered this type of interaction in the databases used. These interactions will not be translated.
Footnotes
| [1] | BioPAX – Biological Pathways Exchange Language - Level 3, Release Version 1 Documentation, 2010. Available: http://www.biopax.org/release/biopax-level3.owl |